Variant Effect Prediction
Empowering Precision Medicine Research
Context
VEP for Precision Medicine
Variant Effect Prediction (VEP) empowers precision medicine by enabling researchers and clinicians to interpret genetic variants and their impacts on protein function, disease mechanisms, and therapeutic targets. WGS and WES analyses identify vast numbers of variants, but understanding their biological significance is challenging. VEP tools help decode these complexities, particularly for:
- Coding Variants: Missense, nonsense, and synonymous variants directly impacting protein structure or function.
- Non-Coding Variants: Regulatory elements like promoters, enhancers, and untranslated regions that influence gene expression.
Yet, VEP is just the starting point. It provides initial predictions, such as identifying a missense mutation, but does not explore deeper questions:
- Where does this variant occur within the protein or regulatory network?
- What functional consequences might arise in specific domains or pathways?
- How can these insights translate to actionable clinical or research outcomes?
How We Can Help
From VEP annotations to translational insights
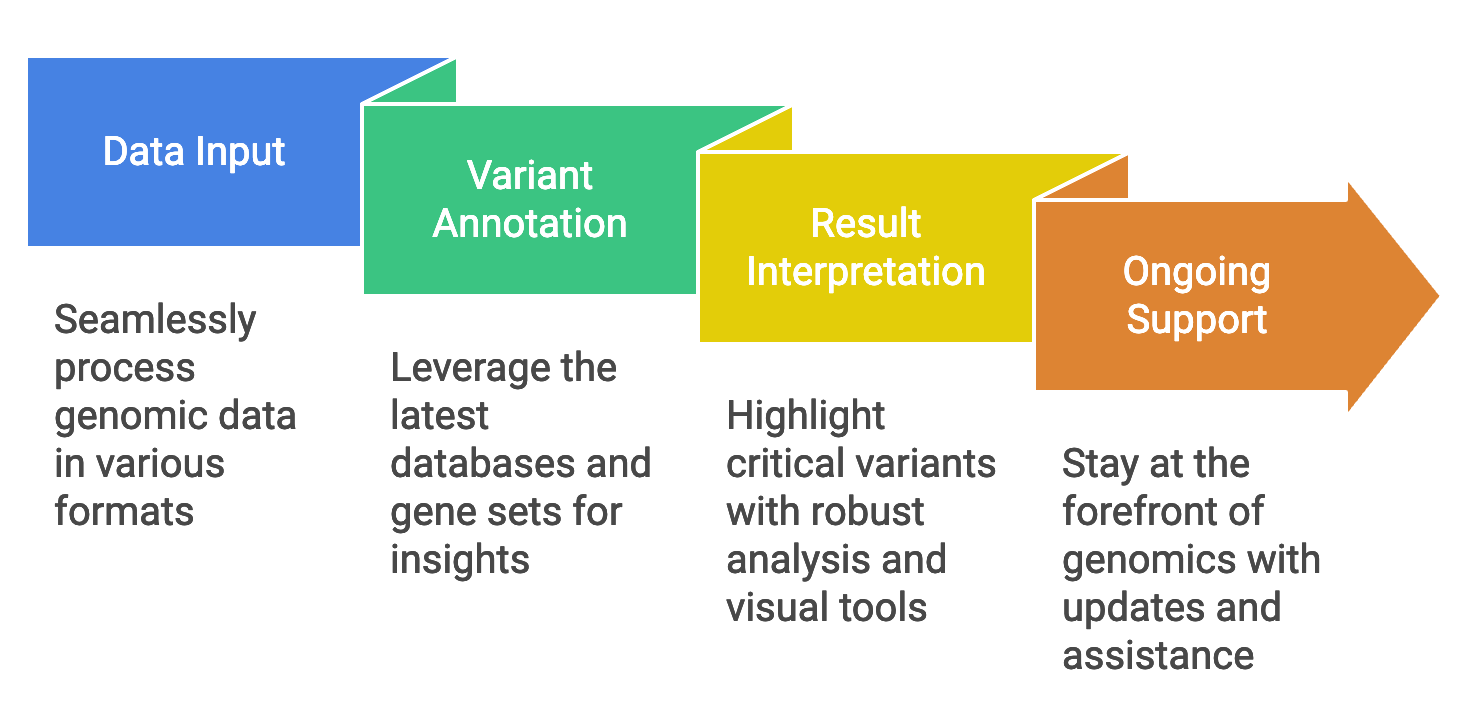
Our advanced VEP services go beyond basic annotations to deliver comprehensive insights and next-step guidance:
- Comprehensive Variant Annotation: Leverage Ensembl VEP to map genomic variants, predict molecular consequences, and provide detailed, actionable insights.
- Advanced Filtering and Prioritization: Customize analyses to focus on clinically relevant variants, enhancing research efficacy and diagnostic accuracy.
- Phenotype Integration: Enrich interpretations by correlating variants with clinical phenotypes, helping uncover causative relationships.
- Coding Variant Analysis: Beyond identifying missense, synonymous, or nonsense mutations, we delve into their location, structural context, and functional impact. For example, a missense mutation in a transporter domain could lead to transport defects—our tools and expertise assess these downstream effects.
- Non-Coding Variant Interpretation: Utilize cutting-edge tools like DNA, codon, and protein-based LLMs to predict impacts on regulatory regions and RNA functions. For example, determining affected genes, even those far from the variant’s position, and assessing changes in transcription, translation, or RNA-based regulation (e.g., microRNAs, silencing RNAs).
- Tool Benchmarking and Evaluation: With over 94 VEP tools available, we assess and rank their performance to help you choose the most reliable for your dataset. For example, benchmarking pathogenicity scores, conservation measures, and functional predictions.
- Multi-Omics Integration: Link genomic variants with gene expression, regulatory evidence, and functional genomic data (e.g., CRISPR models).
- Structural Variant Analysis: Go beyond SNPs to include insertions, deletions, tandem repeats, and inversions for a comprehensive genomic picture.
Highlights
Key components & strengths
High Precision
Advanced algorithms ensure accurate variant classification and functional predictions.
Data Security
Localized tools protect sensitive data, ensuring privacy and compliance.
Comprehensive Coverage
Annotate millions of variants with robust, industry-leading tools like Ensembl VEP.
Scalability
Seamlessly handle small panels or whole-genome datasets, tailored to academic or large-scale research needs.
Customizable Options
Adapt filtering, reporting, and prioritization to meet your specific research goals.
Discover our offerings across the biopharma value chain
Our Solutions
Our Services
Offering services in computational sciences and technology to complement biopharma R&D