Covalent binder design with Gen AI & FBDD
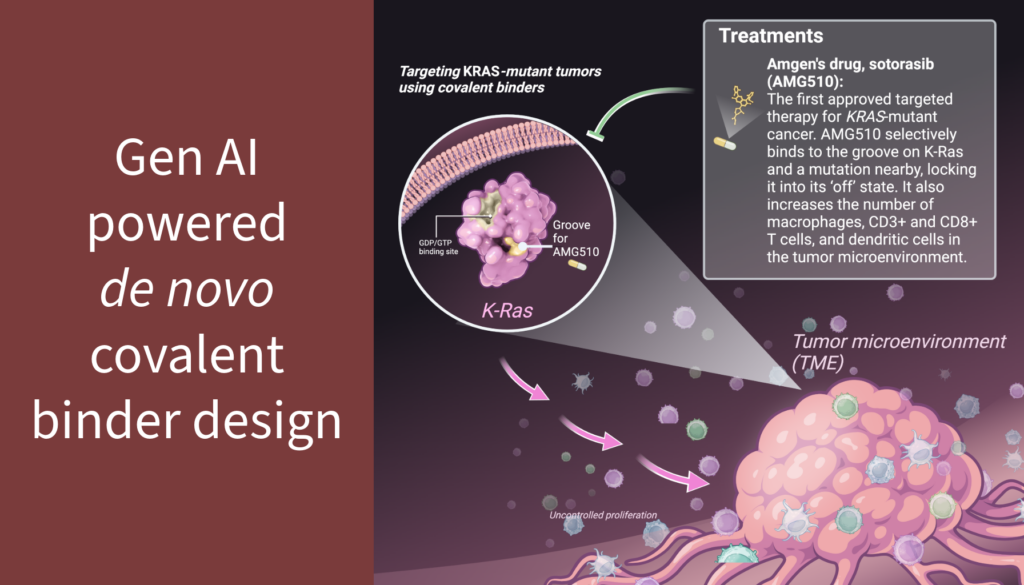
Covalent small molecule drugs are highly valued for their ability to precisely inhibit challenging “hard-to-drug” proteins by forming stable, irreversible bonds with specific residues in the binding pocket. Covalent binding enhances potency, prolongs therapeutic effects, and has proven to be an effective modality for treating cancers.
Design considerations for covalent binders
Designing non-covalent molecules for a given protein pocket involves generating synthesizable, bioavailable compounds that fit tightly within the target pocket. However, designing covalent molecules demands additional considerations. The process begins with identifying a reactive residue in the pocket, followed by designing a compatible warhead capable of forming a covalent bond. The remainder of the molecule must facilitate covalent bond formation while optimizing non-covalent interactions, such as salt bridges, hydrogen bonds, and pi-stacking, with the surrounding residues.
Structure-based drug design (SBDD) methods are not optimized for covalent molecules
The rise of Gen AI and advanced GPU-based algorithms has driven the popularity of SBDD in Biopharma. Over the past decade, numerous computational methods have been developed to accelerate and improve SBDD outcomes. However, these have been primarily focused on non-covalent molecules. For instance, while high-throughput docking of non-covalent molecules has been well-optimized with GPU acceleration, the virtual screening of covalent libraries still needs to be standardized and automated.
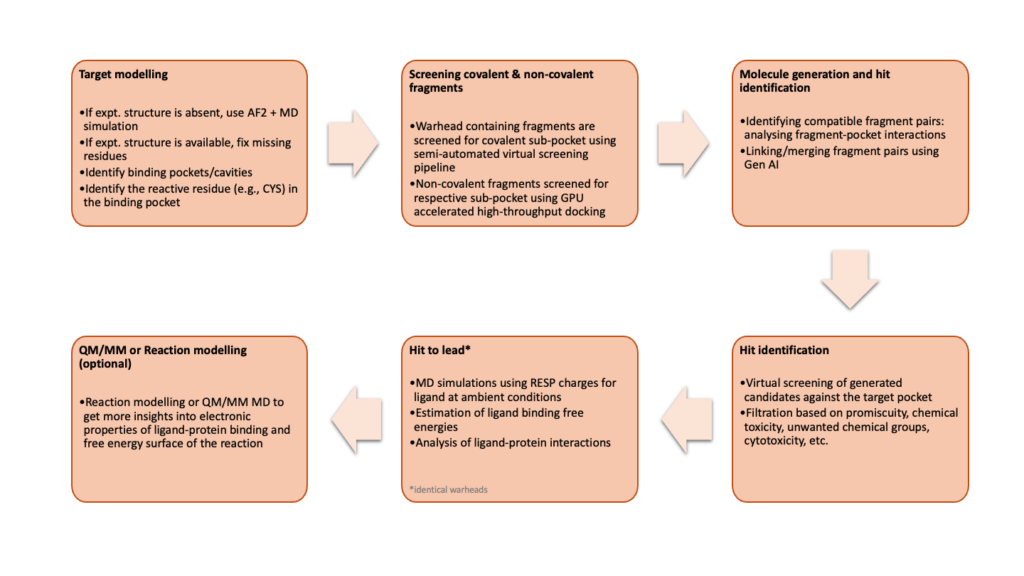
Aganitha’s de novo covalent binder design workflow integrates Gen AI with FBDD
Aganitha has developed a robust in silico workflow for designing de novo covalent binders, specifically targeting cysteine (CYS) residues in proteins. This workflow integrates customizable modules, including protein structure modeling, deep reinforcement learning (RL) with generative AI, and fragment-based drug design (FBDD) (Figure 1). As part of this process, we have optimized several SBDD methods, such as GPU-accelerated high-throughput molecular docking, for covalent molecules.
We can assist you in designing de novo covalent binders tailored to your specific needs. Interested in learning more? Contact us today. To learn more, visit our covalent binders solutions page. Stay tuned for an upcoming case study on designing covalent binders for challenging disease targets.